The objective of this study was to assess the impact of manufacturing-scale, freeze-thaw conditions on aggregation and subvisible particle formation of a monoclonal antibody solution (mAb-A; IgG1) using a small-scale model.
By Kashappa Goud Desai, W. Aaron Pruett, Peter J. Martin, James D. Colandene, Douglas P. Nesta
 Abstract
Abstract
The objective of this study was to assess the impact of manufacturing-scale, freeze-thaw conditions on aggregation and subvisible particle formation of a monoclonal antibody solution (mAb-A; IgG1) using a small-scale model. The temperature-time profiles of manufacturing-scale samples under different freezing and thawing conditions (i.e., slow, medium, and fast freeze-thaw conditions) were generated and used to simulate similar conditions for small-scale samples. Soluble aggregates and subvisible particle counts were measured by size-exclusion chromatography and micro-flow imaging, respectively. Thermal analysis of protein samples was performed by modulated differential scanning calorimetry. The freezing rate in a single freeze-thaw cycle had negligible impact on protein aggregation when fast-thawing conditions were used to thaw. Slow thawing led to higher protein aggregation and subvisible particle formation, which was exacerbated by fast freezing. These effects became more extreme when the number of freeze-thaw cycles was increased from 1 to 3. These trends were found to be similar in large-scale (6.2 L) and small-scale (30 mL and 100 mL) assessments, with the total magnitude of degradation higher in the small-scale system. The systematic small-scale model employed in the current study was predictive of manufacturing-scale freeze-thaw conditions.
Therapeutic proteins, peptides, and antibodies have emerged as highly effective modern medicines for numerous diseases and disorders (1). Therapeutic monoclonal antibodies (mAbs) represent a rapidly growing area in the biopharmaceutical sector (2). Enormous success achieved with mAbs can be attributed to their distinctive beneficial properties and advantages, which include high binding specificity and affinity, availability of humanized forms that can attenuate immunogenic responses, and robust manufacturing processes (3, 4). Poor stability of monoclonal antibodies outside their natural environment, however, is one of the major challenges in product development (2–6). A number of non-optimal formulation, manufacturing, or storage conditions often cause instability of mAbs, which in turn can affect the bioactivity of proteins (7).
Freezing and thawing are integral steps in the manufacturing of most biopharmaceutical products. Storage of biopharmaceuticals in the frozen state has distinctive advantages. It minimizes the risk of microbial growth, increases product stability with extended shelf life, eliminates agitation and foaming during transportation, and increases flexibility during manufacturing (7–11). Freezing and thawing stresses (e.g., cold-denaturation, cryo-concentration, ice formation, crystallization of buffer or non-buffer components, phase separation, redistribution of solutes, pH fluctuation, and thawing time), however, can induce complex physical and chemical changes in the solvent/solute conditions, which in turn can potentially lead to denaturation and aggregation of proteins (10, 12-16).
Formation of protein aggregates and subvisible particles during the manufacturing of drug products are a major concern due to the potential immunogenicity of protein aggregates in patients (17–22). There have been studies to determine the impact of potential freeze-thaw factors on aggregation of proteins. A number of potential formulation variables (e.g., buffer composition, pH, ionic strength, and cryoprotectants) and freeze-thaw stresses (e.g., crystallization of excipients, ice formation, and freeze concentration) have been evaluated (12, 15, 19, 23-35). There is also a growing interest in the pharmaceutical industry to develop small-scale models to study the potential impact of large-scale freeze-thaw process variables on protein stability. The small-scale models are cost-effective and less time consuming (36). A decision regarding freeze-thaw parameters must often be made prior to having sufficient amounts of the product available to assess process characterization at manufacturing scale, thereby necessitating use of small-scale studies.
One approach to scale-down freeze-thaw process characterization studies is to use a temperature-controlled chamber to expose small samples of a formulation to time-temperature profiles that simulate and/or bracket that of the manufacturing-scale process. This approach offers an improvement over uncontrolled freeze-thaw of small samples, which tend to occur quickly and may suppress stochastic phenomena such as ice nucleation or kinetic phenomena such as full crystallization of eutectic phases (37). A small-scale version of controlled freezing-thawing systems is useful for scale-down assessment of freeze-thaw process characterization (37). The authors, therefore, sought to assess the feasibility of a small-scale model to determine the impact of large-scale freeze-thaw conditions on aggregation and subvisible particle formation of a monoclonal antibody (mAb, IgG1).
MATERIALS AND METHODS
Monoclonal antibody (mAb-A, IgG1) formulation
The mAb-A formulation (64 mg/mL protein concentration) was prepared in a formulation that contained a buffering agent (10 mM), surfactant, cryoprotectant (disaccharide) and bulking agent (glycine). The formulation samples were filtered through a 0.22 µm filter into ~8.3 L, 100 mL, and 30 mL bags (Sartorius Stedim) with thermowells. Excess air was purged from the bags to minimize air-liquid interfaces. A thermal surrogate solution (64 mg/mL of disaccharide in formulation buffer) was used to minimize the amount of protein solution needed to add thermal mass for the large-scale experiments and reach different freezing profiles. Custom designed stainless-steel cases lined with foam padding (designed to protect the bags while in the frozen state) were used as containers for the ~8.3 L bags during storage and handling.
Size-exclusion chromatography (SEC)
Soluble aggregates in protein samples were measured using a high-performance liquid chromatography (HPLC) system attached with a 7.8 mm × 30 cm column (Tosoh BioScience). Approximately 350 µg protein was loaded onto the column. The detection wavelength was 280 nm. The column equilibration (65 minutes) and sample elution (35 minutes) was done with the mobile phase (50 mM sodium citrate, 450 mM sodium chloride, pH 6.5) run at 0.5 mL/min. Monomer, aggregate, and low-molecular weight species levels were calculated as a percentage of the total protein peak areas. Only aggregate levels are reported here because low-molecular weight species were unchanged in all conditions. Based on the qualified intermediate precision of the method, changes in percent aggregate greater than 0.1% were considered significant.
Micro-flow imaging (MFI)
The concentration of subvisible particles in 1-100 µm size range was measured by a micro-flow imaging (MFI) system (Brightwell) equipped with a 100 µm/1.6 mm flow cell. The measurement involved flushing of the flow cell with a 0.25 mL sample, followed by imaging analysis of a 0.65 mL sample. Reported values are the average of two samples, with an acceptance criterion of <30% difference between the two values.
Low-temperature differential scanning calorimetry (mDSC)
Thermal events were measured in frozen solutions using a low-temperature differential scanning calorimeter (mDSC) (TA Instruments). A mechanical cooling accessory (RCS90) was used for cooling the sample chamber to temperatures as low as -80 ˚C. Dry nitrogen was used as a purge gas at a flow rate of 50 mL/min. A volume of 20 µL of solution was placed in an aluminum sample pan with a capacity of 40 µL. An aluminum lid was placed on the sample pan and was hermetically sealed using a crimping press. An empty aluminum pan with lid, identical to that used for the sample, was used as the reference. The sample and reference pans were cooled at a controlled rate of 5 ˚C per minute for fast freezing conditions and equilibrated at the lowest programmed temperature for 10 minutes. The slow freeze rate was at a controlled rate of 0.1 ˚C per minute, and sample and reference pans were equilibrated at the lowest programmed temperature. The modulation amplitude was adjusted to allow cooling during temperature modulation to amplify weak heat capacity signals. The melting and crystallization events are reported at the onset of the thermal event using the non-reversing signal. A smoothing region width of 1.000 ˚C was applied to the non-reversing heat flow signal.
Large-scale freeze-thaw cycling
Approximately 6.2 L mAb-A formulation were filled into 8.3 L bags (Sartorius Stedim). Prior to freezing, the bags were stored at 2-8 ˚C, and once frozen, they were stored at -80 ˚C. Modified freezing and thawing cycles were designed and used to achieve a broad range of temperature profiles (Table I). The freezing was conducted in -80 ˚C freezers (bags reached ≤70 ˚C), which were underpowered for freezing large volumes of aqueous solutions, causing the freezing temperature profiles to be load-dependent. Surrogate sample bags were utilized for adding thermal mass for the large-scale freezing experiments at -80 ˚C, to ensure reliability of freezing temperature profiles. The temperature profiles were recorded with multiple type-T thermocouples placed in the thermowells and attached to the upper and lower surface (Figure 1). The endpoint of the thaw was complete melting of visible ice. At the end of each thaw, the bags were mixed by multiple inversions. The bags were exposed from 1 to 3 cycles (1x-3x) of either fast freeze/fast thaw, fast freeze/slow thaw, medium freeze/medium thaw, slow freeze/fast thaw, or slow freeze/slow thaw. A liquid control was placed at 2-8 ˚C during the entire duration of the study. A -80 ˚C control was also generated (this was exposed to one medium freeze-medium thaw cycle). Samples from each test condition were taken after the first and third freeze/thaw cycle (1x and 3x), and analyzed by size-exclusion chromatography (SEC) (along with the control samples). The 3x freeze-thaw samples were also analyzed for subvisible particles by MFI.
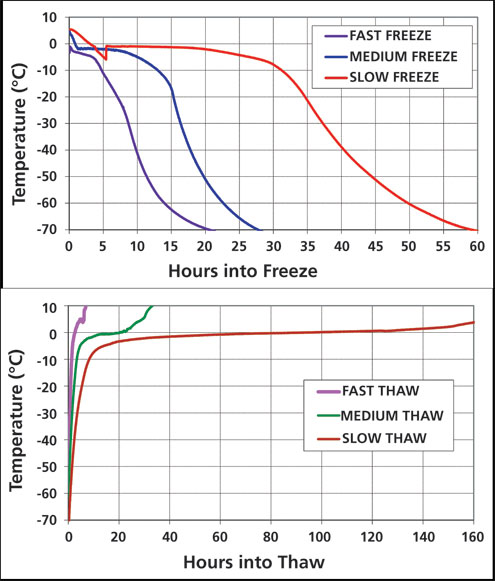
Figure 1: Temperature profiles based on average of four thermocouples for freezing (top) and thawing (bottom) of 6.2 L aliquots of mAb-A in 8.3 L bags.
Scale-down freeze-thaw simulation
Approximately 30 mL and 100 mL of 64 mg/mL mAb-A were filled into 30mL and 100mL bags (Sartorius Stedim), respectively. The 6.2 L fill was a convenient volume (6-7 L) that manufacturing used for BDS (logistically ~6-7 L per bag was a suitable volume). Regardless, the surface area to volume ratio is higher in the 100 mL and 30 mL bags. Scale-down process simulation was done by using a programmable temperature control system (Sartorius Stedim) to apply a series of linear temperature ramping profiles to a sample chamber. The bags were installed in the chamber in such a manner that both side edges were in contact with the heat transfer surfaces. The sample chamber was insulated during operation to minimize environmental heat transfer.
For each large-scale freeze-thaw cycle (6.2 L in 8.3 L bag), the four thermocouple trends were averaged to generate a target freezing and thawing temperature profile (Figure 1), and a scale-down simulation program was developed to mimic each manufacturing-scale profile (a separate profile with custom-made temperature-time steps to mimic each manufacturing-scale profile was created and used to run small-scale processes via a custom-built software from Sartorius). The cycles were adjusted for heat loss in the control system. Supercooling steps to induce ice nucleation were added only for the medium and slow freezing rates. Time required for freezing to ≤ -40 ˚C ranged from 10-40 hours. The time required for completion of thawing, as indicated by thermocouple rising above 0 ˚C, ranged from approximately 3-125 hours. Samples subjected to the scale-down simulation were analyzed by SEC (after 1x and 3x freeze/thaw cycles). The 3x samples were also analyzed for subvisible particles by MFI.
RESULTS AND DISCUSSION
Effect of freeze-thaw process variables on aggregation and subvisible particle formation of mAb-A
Robust freezing and thawing processes are essential for manufacturing and storage of biopharmaceutical products. Optimal conditions ensure improved protein stability during these stages. Logistically, small-scale models are ideal to assess the impact of freeze-thaw process variables on protein stability. Small-scale models, however, may not reflect the actual freezing or thawing rates, or the effect on product, that occur at manufacturing scale. Small-scale freeze-thaw systems have been introduced that are capable of more accurately mimicking freezing and thawing rates that occur at manufacturing scale (37). Using these systems, the large-scale freezing and thawing environments can be simulated in small-scale models. In this study, the authors sought to evaluate and compare the impact of freeze-thaw process variables using both large-scale and small-scale models.
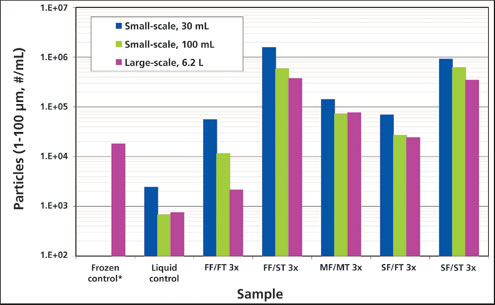
Large-scale freeze-thaw temperature profiles were successfully simulated in scale-down studies using a freeze-thaw unit (Sartorius Stedim) (applied programmed temperature profiles to freeze and thaw 30 mL and 100 mL bags) (Figure 1). The impact of freeze-thaw process parameters (e.g., freezing and thawing rates, number of cycles) on protein aggregation, as generated from the small-scale and large-scale models, is shown in Figure 2. A frozen control had a similar level of aggregate to that of the 1x medium freeze-medium thaw sample, indicating that the storage parameter alone did not influence the aggregation of mAb A. For a single freeze-thaw cycle, the freezing rate had a negligible impact on protein aggregation when followed by fast thawing conditions (percent aggregates for 1x slow, medium, and fast-freeze samples were ≤ 0.1% different, relative to that observed for the liquid control), and was similar for both large-scale and small-scale sample sizes. Slow thawing, however, led to significantly higher protein aggregation (percent aggregates for 1x slow-thaw and medium-thaw samples of small-scale and large-scale studies were >0.1% different when compared with the liquid control or the 1x fast-thaw samples). Interestingly, the negative impact of slow thawing appeared to be exacerbated by fast freezing, as this condition generated the highest levels of aggregates. Conversely, slow freezing followed by fast thawing had the lowest levels of aggregate formation.
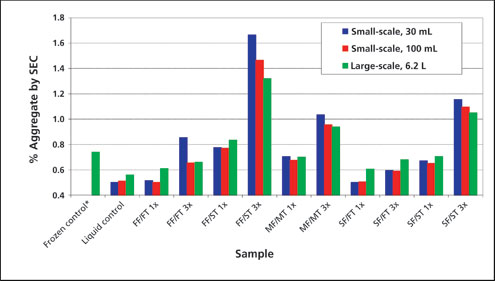
The aggregation of mAb-A was impacted by the number of freeze-thaw cycles under all test conditions (3x samples had higher aggregate levels compared with corresponding 1x samples). Among all the 3x samples, 30mL fast freeze/slow thaw sample had highest level of aggregates. After 3x fast freeze/slow thaw cycles, the aggregate level in different size samples was found to be in the following order: 30mL sample size >100mL sample size >6.2L. Moreover, the trends observed with respect to freezing and thawing conditions were the same regardless of the scale.
Representative (3x fast-freeze/fast-thaw, fast-freeze/slow-thaw, medium-freeze/medium-thaw, slow-freeze/fast-thaw, and slow-freeze/slow-thaw samples) SEC chromatograms from the study are shown in Figure 3. These overlays show that the soluble aggregates are predominantly comprised of dimers. Due to limitations in the analytical capability of the SEC method used, and the relatively low aggregate level, it was not possible to quantitatively assign the percent of dimer versus higher-order multimers.
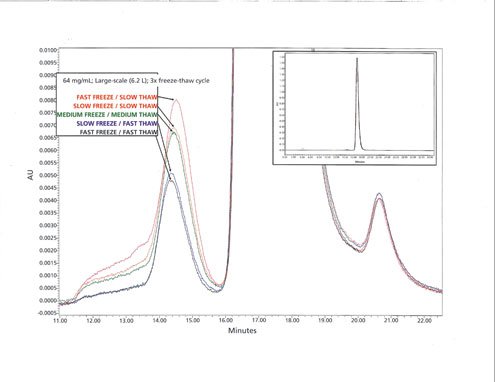
Figure 3: Size-exclusion chromatography (SEC) chromatogram of 3x fast-freeze/fast-thaw, fast-freeze/slow-thaw, medium-freeze/mediumthaw, slow-freeze/fast-thaw and slow-freeze/slow-thaw samples.
Aggregation of proteins during freeze-thaw cycling has generally been attributed to partial unfolding of protein molecules caused by the perturbing conditions such as pH variation, low temperature, freeze concentration of solutes, exposure of proteins to ice-liquid interface or surfaces induced by excipient crystallization, and/or adsorption to materials of contact (8, 10, 13, 16, 28, 33, 35). The formulation buffer used in the current study contains glycine as a bulking agent. It is known that glycine may crystallize under frozen storage conditions and form a new surface, thereby potentially causing protein denaturation (27, 31). During thawing, recrystallization can cause additional protein perturbations at the ice-liquid interface (38). The cause for higher protein unfolding under slow-thawing conditions has been linked to prolonged exposure of protein to low temperature and high solute concentration medium (39). The findings of the current study were in good agreement with those reported by Cao et al. (38), wherein slow thawing conditions caused higher protein degradation and loss of activity (38).
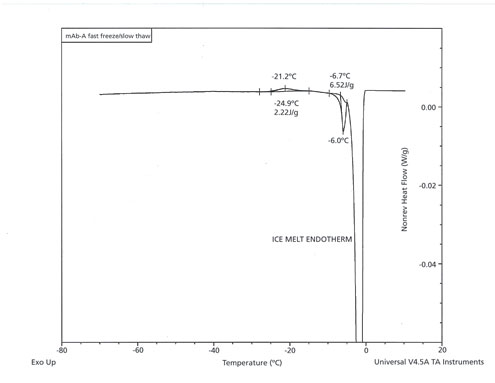
One key finding from this work is that fast freezing followed by slow thawing resulted in greater degradation of the mAb than did slow freezing followed by slow thawing. If glycine crystallization is the primary cause of aggregation, it would suggest that the crystallization of glycine during thawing is more detrimental than the crystallization of glycine during the freezing step. This finding is assumed because rapid freezing would be more likely to leave glycine in the amorphous phase after the freeze compared with slow freezing. It would then follow that rapid freezing followed by slow thawing would be more likely to lead to higher levels of glycine crystallization during the thaw. The authors sought to study this phenomenon by mDSC. The findings of mDSC were found to be in good agreement with this hypothesis. In the DSC experiment, where fast freezing was followed by slow thawing, an exothermic peak at approximately -25 °C onset, likely corresponding to glycine (eutectic) crystallization, followed by a likely eutectic melt around 6.6 °C onset, were readily observed during the slow thawing step (Figure 5). Neither the exothermic thermal event nor the endothermic melt were observed after fast freezing and fast thawing conditions (data not shown).
It can be summarized that fast freezing and fast thawing conditions are optimal for mAb-A to prevent potential effects such as protein aggregation and subvisible particle formation, but that the fast thawing rate is the more important parameter. The small-scale model successfully identified the impact of manufacturing-scale freezing and thawing rates on aggregation and subvisible particle formation of a mAb-A, thereby suggesting its feasibility and benefits in biopharmaceutical manufacturing. The simulation at small-scale amplified the affects observed at large-scale possibly due to difference in interfacial contact effects (e.g., ice crystal, air, and container surface).
CONCLUSION
A robust manufacturing-scale, freeze-thaw process suitable for mAb-A was developed through large-scale and small-scale assessment models. A small-scale process simulation model evaluated in the current study successfully identified risk at manufacturing-scale conditions on aggregation and subvisible particle formation of mAb-A. The study also demonstrated the importance of thawing parameters. Slow thawing was shown to have the most negative impact on the product (highest levels of aggregate formation). Fast freeze-fast thaw conditions were found to be optimal for mAb-A. Multiple freeze-thaw cycles are sometimes inevitable during manufacturing due to unforeseeable issues. Multiple cycles under fast-freeze/fast-thaw conditions are also considered to be feasible for mAb-A.
ACKNOWLEDGEMENT
The authors thank Dr. Melissa D. Perkins, PhD currently at the Department of Manufacturing Science and Technology, Hospira, USA for her feedback.
REFERENCES
1. T.K. Das, AAPS PharmSciTech. 13 (2), pp. 732-746 (2012).
2. M. Vázquez-Rey, D.A. Lang, Biotechnol. Bioeng. 108 (7), pp. 1494-1508 (2011).
3. A. Beck et al., Nat. Rev. Immunol. 10 (5), pp. 345-352 (2010).
4. P. Kheddo et al., Int. J. Pharm. 473 (1-2), pp. 126-133 (2014).
5. V. Rombach-Riegraf et al., PLoS ONE. 9 (1), e86322 (2014).
6. H.C. Mahler et al., J. Pharm. Sci. 98 (9), pp. 2909-2934 (2009).
7. P.A. Shamlou et al., Biotechnol. Appl. Biochem. 46 (1), pp. 13-26 (2007).
8. K. Ho et al., Am. Pharm. Rev. 11 (5), pp. 64-70 (2008).
9. P. Kolhe et al., BioPharm Int. 23 (6), pp. 53-60 (2010).
10. S.K. Singh et al., Pharm. Res. 28 (4), pp. 873-885 (2011).
11. S.K. Singh, Am. Pharm. Rev. 10 (3), pp. 26-33 (2007).
12. B.S. Bhatnagar et al., J. Pharm. Sci. 97 (2), pp. 798-814 (2008).
13. L.A. Kueltzo et al., J. Pharm. Sci. 97 (5), pp. 1801-1812 (2008).
14. L. Liu et al., J. Pharm. Sci. 103 (7), pp. 1979-1986 (2014).
15. W. Liu et al., AAPS PharmSciTech. 6 (2), pp. E150-E157 (2005).
16. S.K. Singh et al., BioProcess Int. 7 (10), pp. 34-42 (2009).
17. F. Baert et al., New Engl. J. Med. 348 (7), pp. 601-608 (2003).
18. V. Filipe et al., mAbs 4 (6), pp. 740-752 (2012).
19. A. Hawe et al., Eur. J. Pharm. Sci. 38 (2), pp. 79-87 (2009).
20. S. Hermeling et al., Pharm. Res. 21 (6), pp. 897-903 (2004).
21. A.S. Rosenberg, AAPS J. 8 (3), pp. E501-E507 (2006).
22. R.L. West et al., Aliment Pharmacol. Ther. 28 (9), pp. 1122-1126 (2008).
23. P. Sundaramurthi et al., Pharm. Res. 27 (11), pp. 2374-2383 (2010).
24. U.H. Verkerk et al., J. Mass. Spectrom. 38 (6), pp. 618-631 (2003).
25. S. Vemuri et al., PDA J. Pharm. Sci. Technol. 48 (5), pp. 241-246 (1994).
26. M.A. Rodrigues et al., J. Pharm. Sci. 100 (4), pp. 1316-1329 (2011).
27. K.A. Pikal-Cleland et al., J. Pharm. Sci. 91 (9), pp. 1969-1979 (2002).
28. J.M. Sarciaux et al., J. Pharm. Sci. 88 (12), pp. 1354-1361 (1999).
29. S. Saito et al., Pharm. Res. 30 (5), pp. 1263-1280 (2013).
30. B.A. Kerwin et al., J. Pharm. Sci. 87 (9), pp. 1062-1068 (1998).
31. D.B. Varshney et al., Pharm. Res. 24 (3), pp. 593-604 (2007).
32. P. Kolhe et al., Biotechnol. Progr. 26 (3), pp. 727-733 (2010).
33. R. Zhou et al., PDA J. Pharm. Sci. Technol. 66 (3), pp. 221-235 (2012).
34. X. Li, S.L. Nail, J. Pharm. Sci. 94 (3), pp. 625-631 (2005).
35. G.B. Strambini, E. Gabellieri, Biophys. J. 70 (21), pp. 971-976 (1996).
36. M. Puri et al., BioProcess Int. 13 (1), pp. 34-45 (2015).
37. C. Padala et al., PDA J. Pharm. Sci. Tech. 64 (4), pp. 290-298 (2010).
38. E. Cao et al., Biotechnol. Bioeng. 82 (6), pp. 684-690 (2003).
39. K.A. Pikal-Cleland et al., Arch. Biochem. Biophys. 384 (2), pp. 398-406 (2000).